Products High performance NGS
Genalice provides a modular high performance NGS secondary analysis suite.
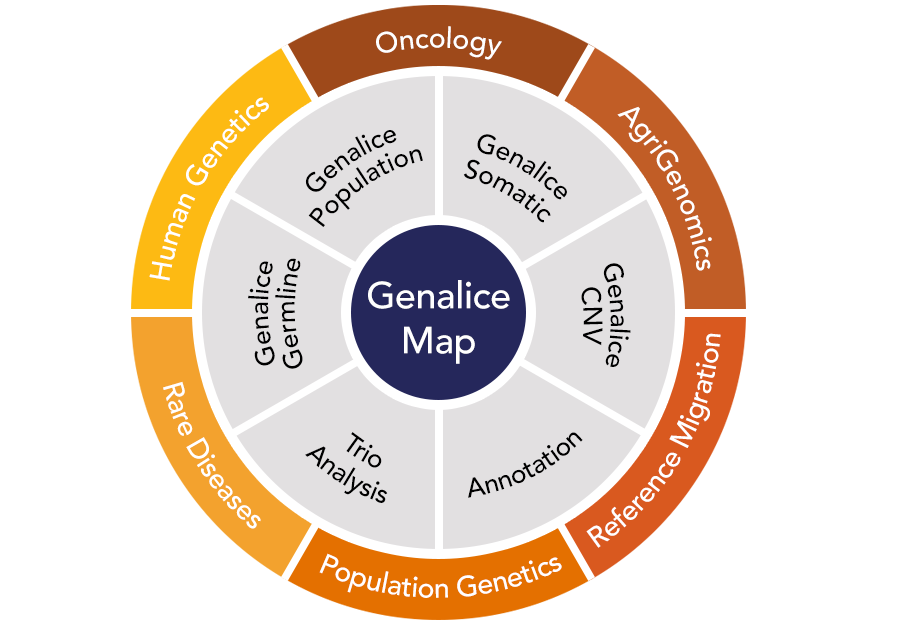
Genalice creates a reliable bridge between NGS data streaming from the sequencer (FASTQ or unaligned BAM) to detailed genetic interpretation and reporting solutions.
The patented data processing methods, the rigorous dataflow implementation, the scalability of the software architecture and the efficiency of Genalice data formats are specifically designed for high volume NGS data management. Together it allows to do cost effective and timely heavy data lifting, which is unmet in the market.
Genalice solutions can be deployed at any scale desired for secondary analysis of NGS data and leads to near time diagnosis, treatment selection and biomarker discovery.
The benefits of Genalice solutions
SPEED
From FASTQ to VCF in less than 29 minutes.

ACCURACY
Highly accurate and precise alignment and variant calling.

EASE OF USE
All-in-one solution simple workflow and flexible deployment.

FILESIZE
Twentyfold storage footprint reduction of aligned read files.
There are no products listed under this category.
